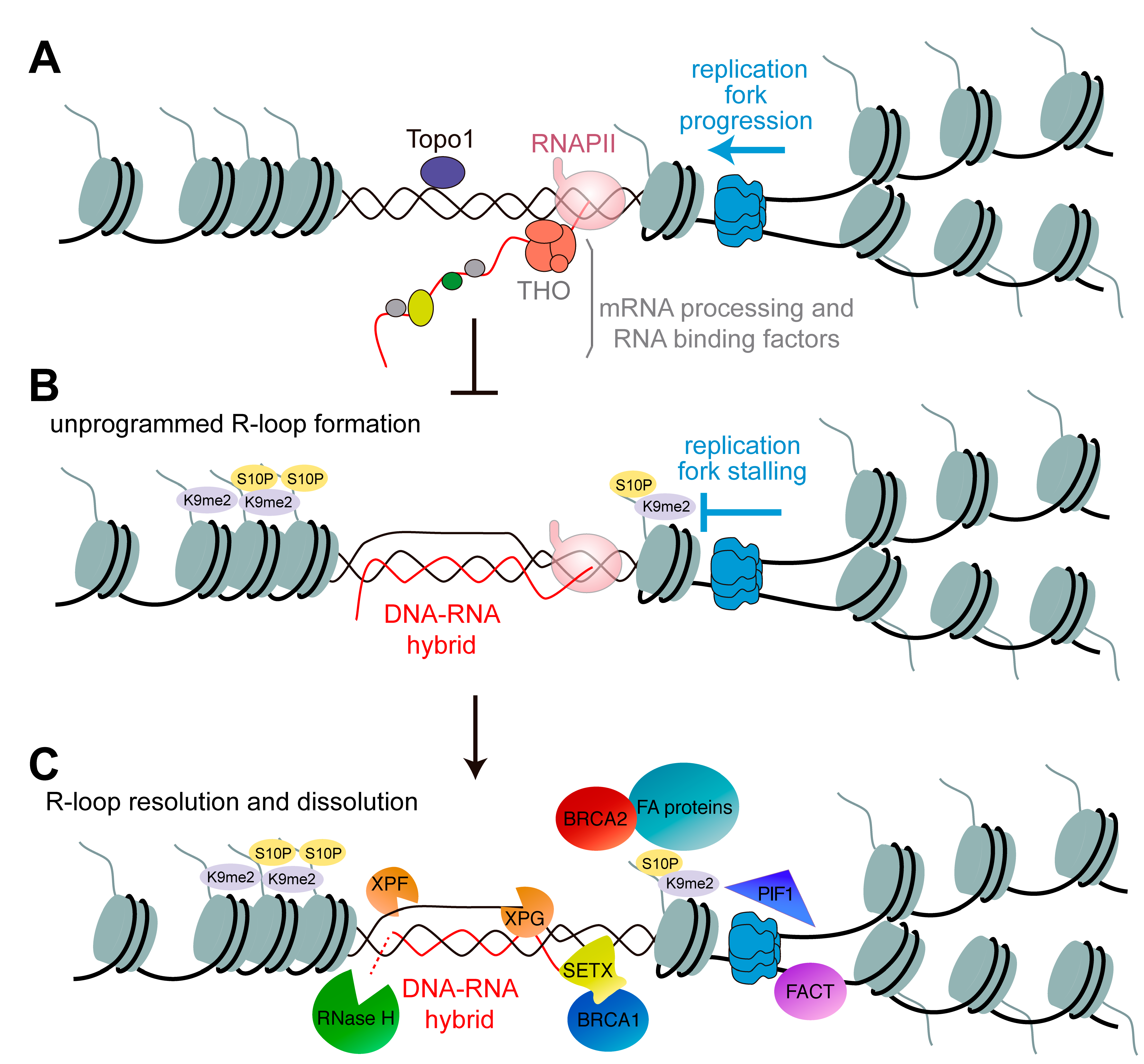
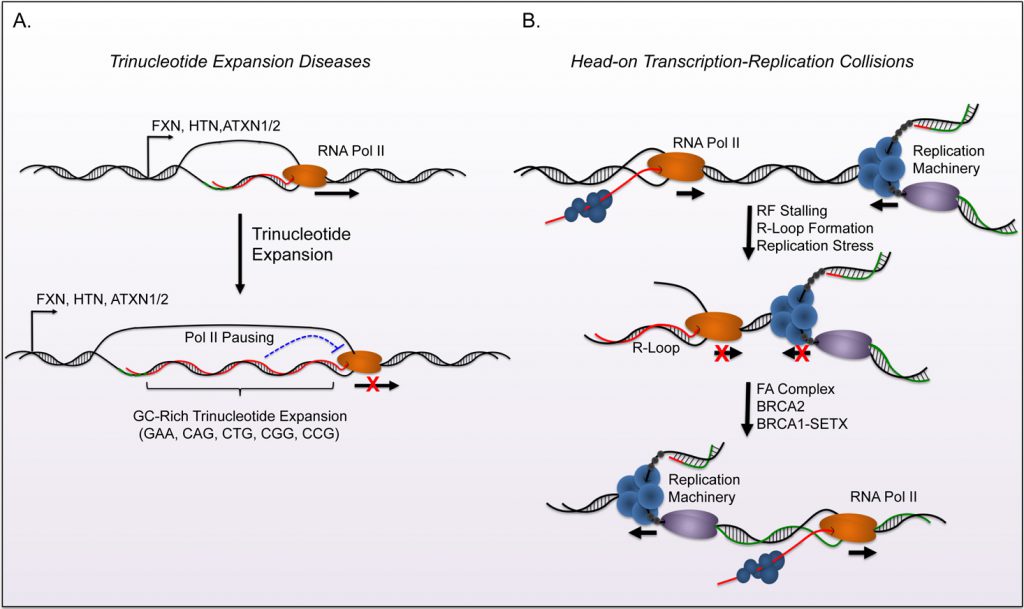
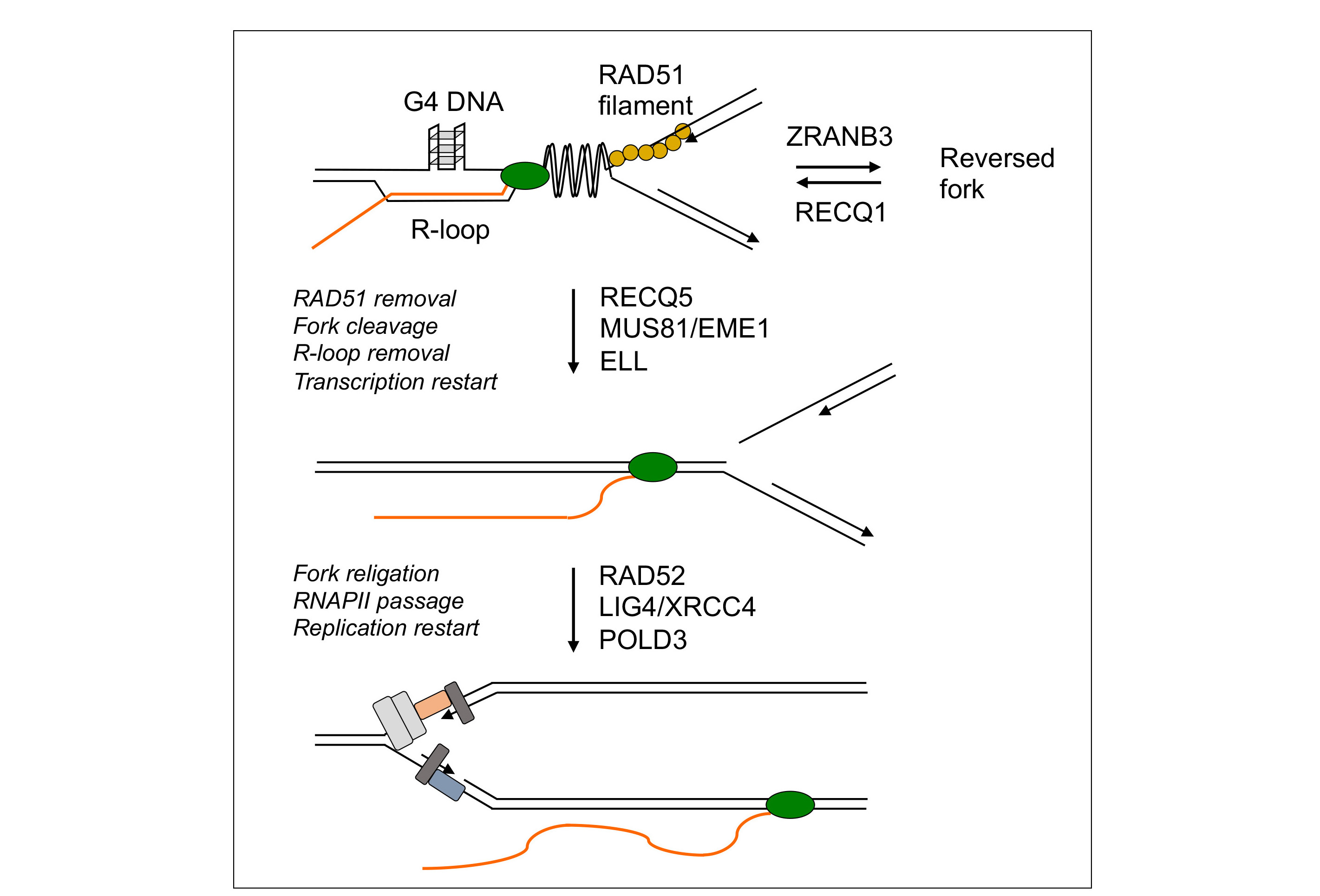
Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis - ScienceDirect
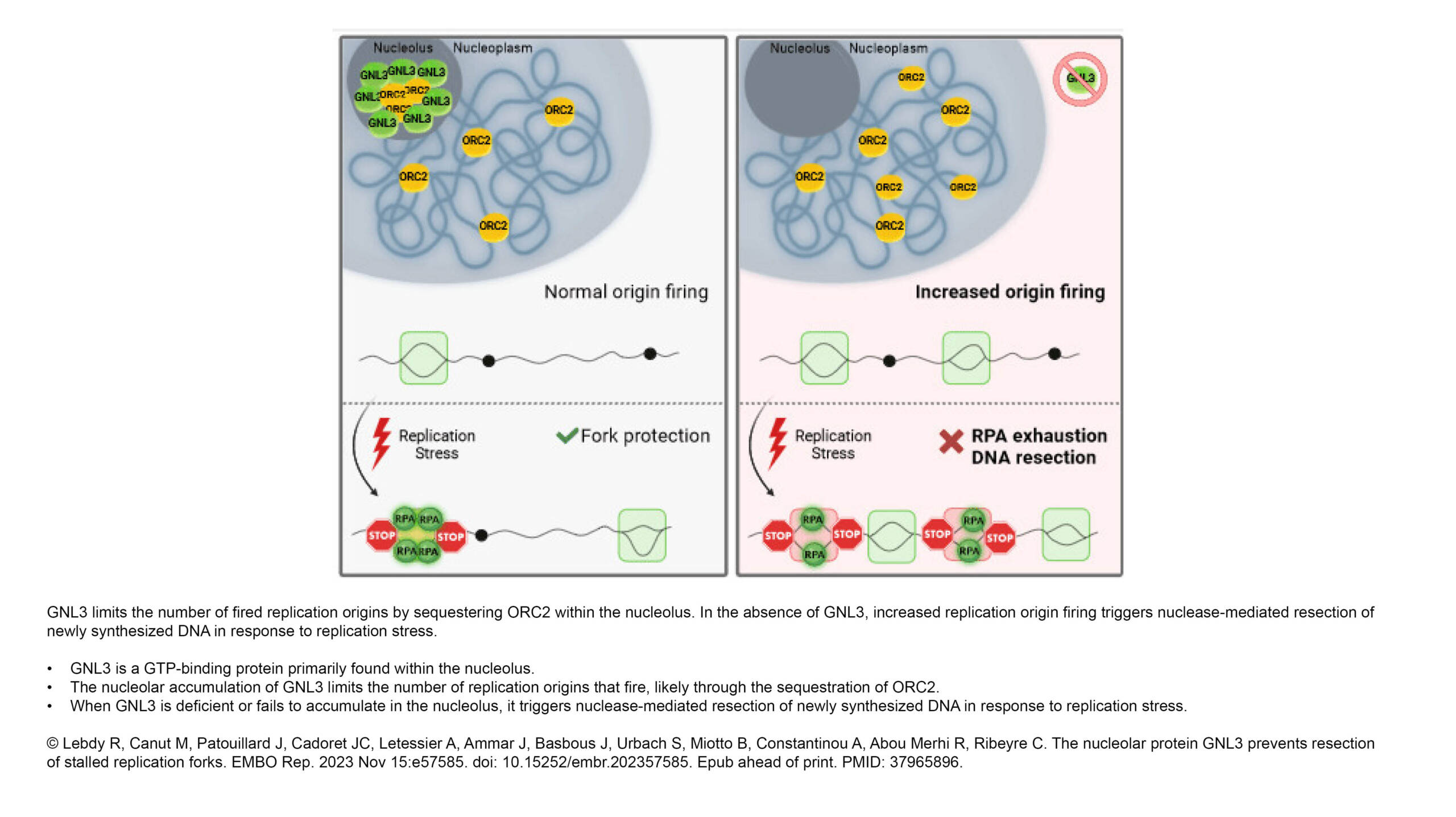
Cadoret Lab - The nucleolar protein GNL3 prevents resection of stalled replication forks - Institut Jacques Monod

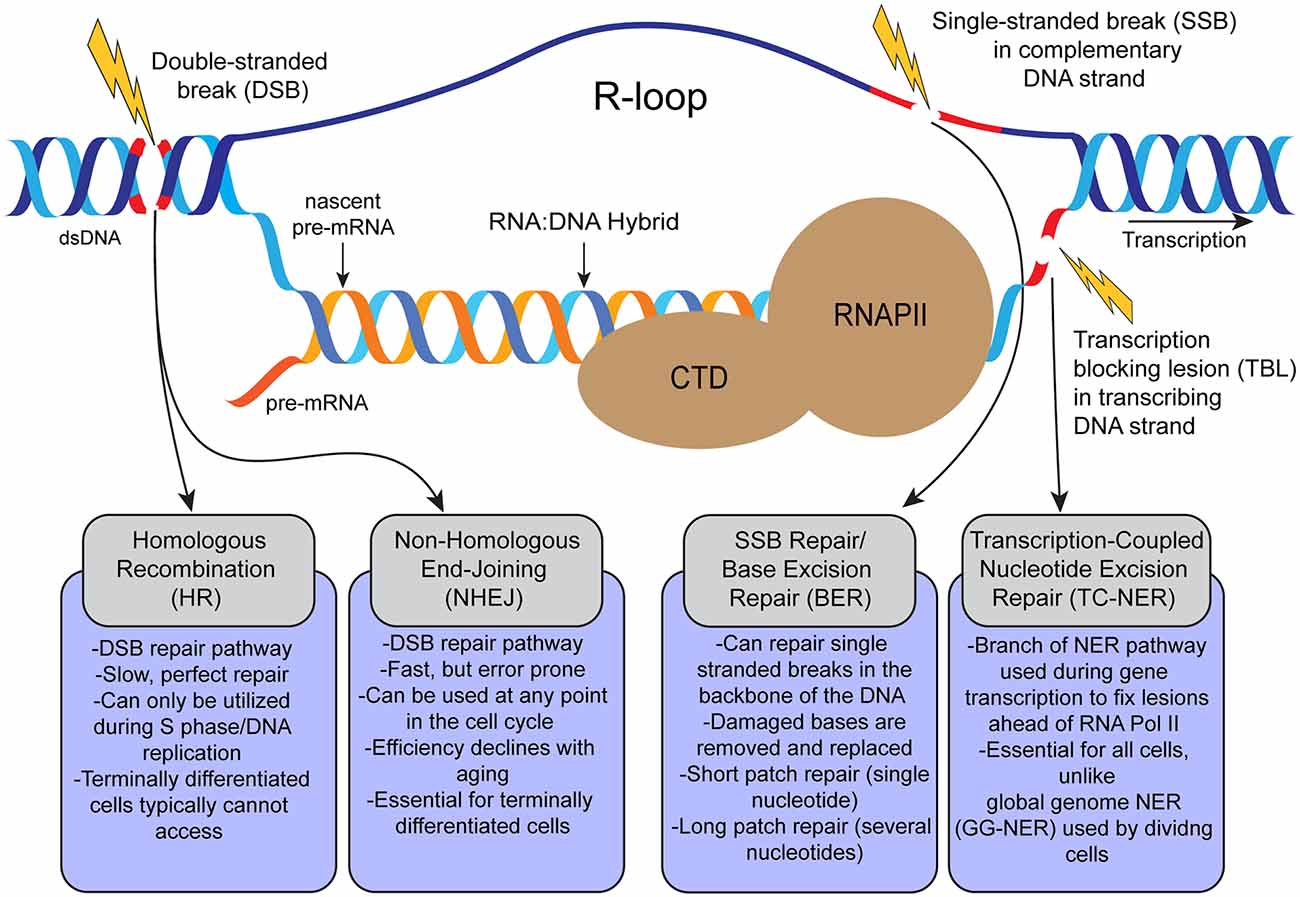
RNase H eliminates R‐loops that disrupt DNA replication but is nonessential for efficient DSB repair | EMBO reports
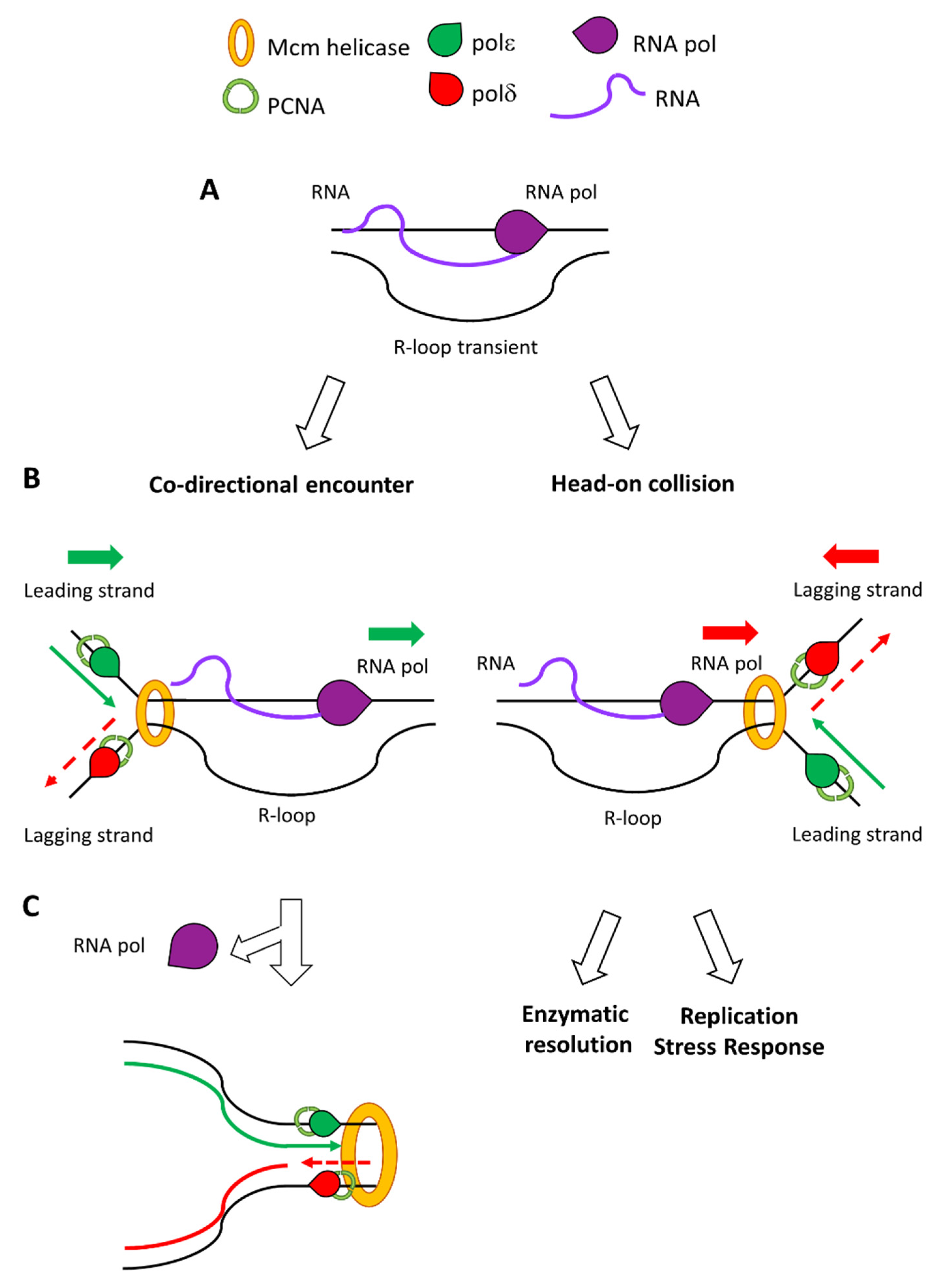
IJMS | Free Full-Text | From R-Loops to G-Quadruplexes: Emerging New Threats for the Replication Fork
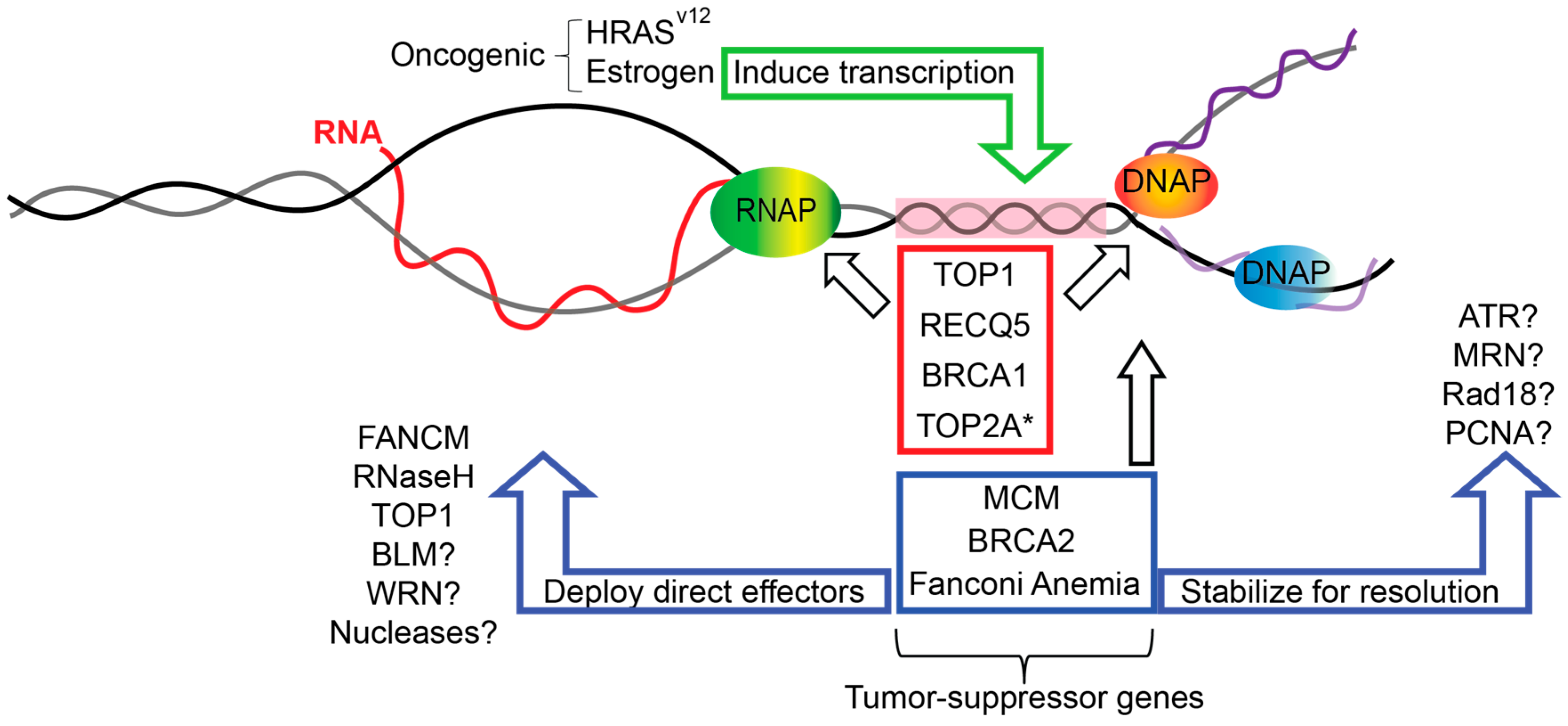
Genes | Free Full-Text | Replication Fork Protection Factors Controlling R-Loop Bypass and Suppression

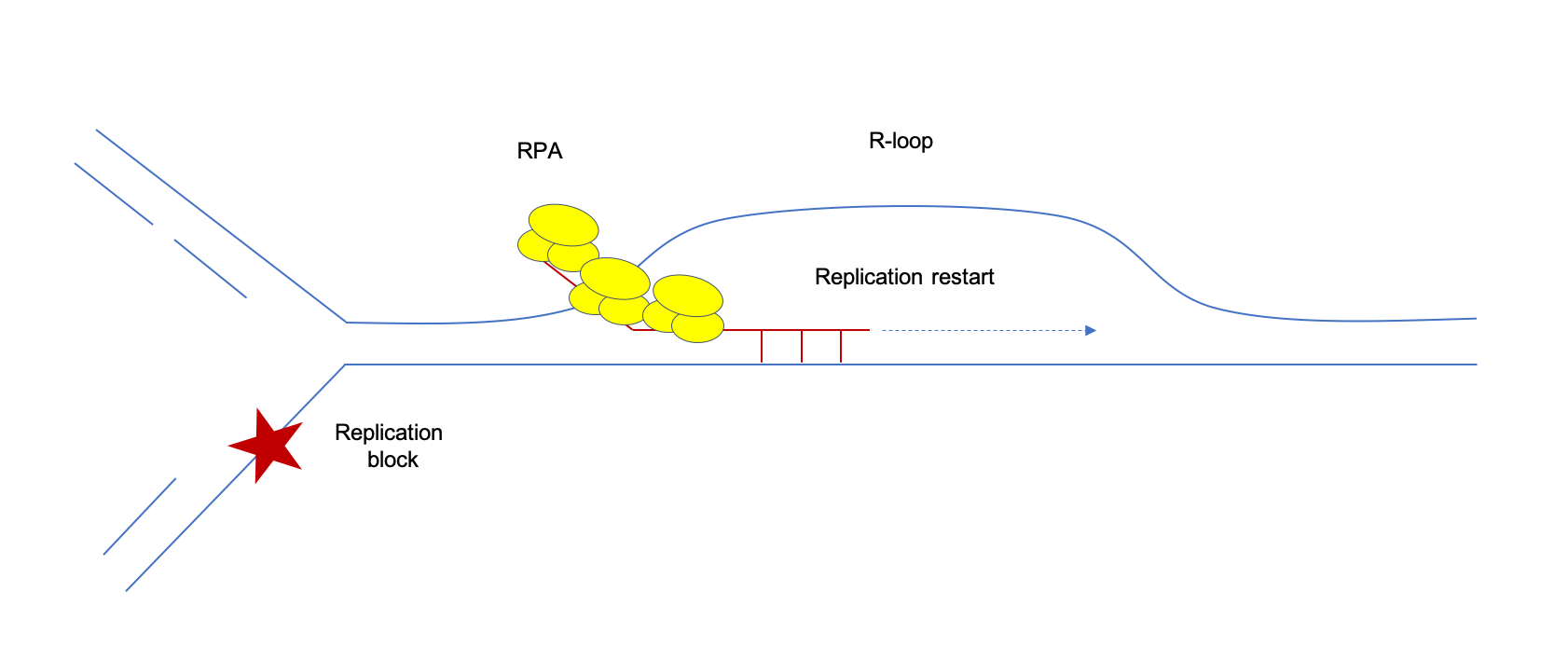
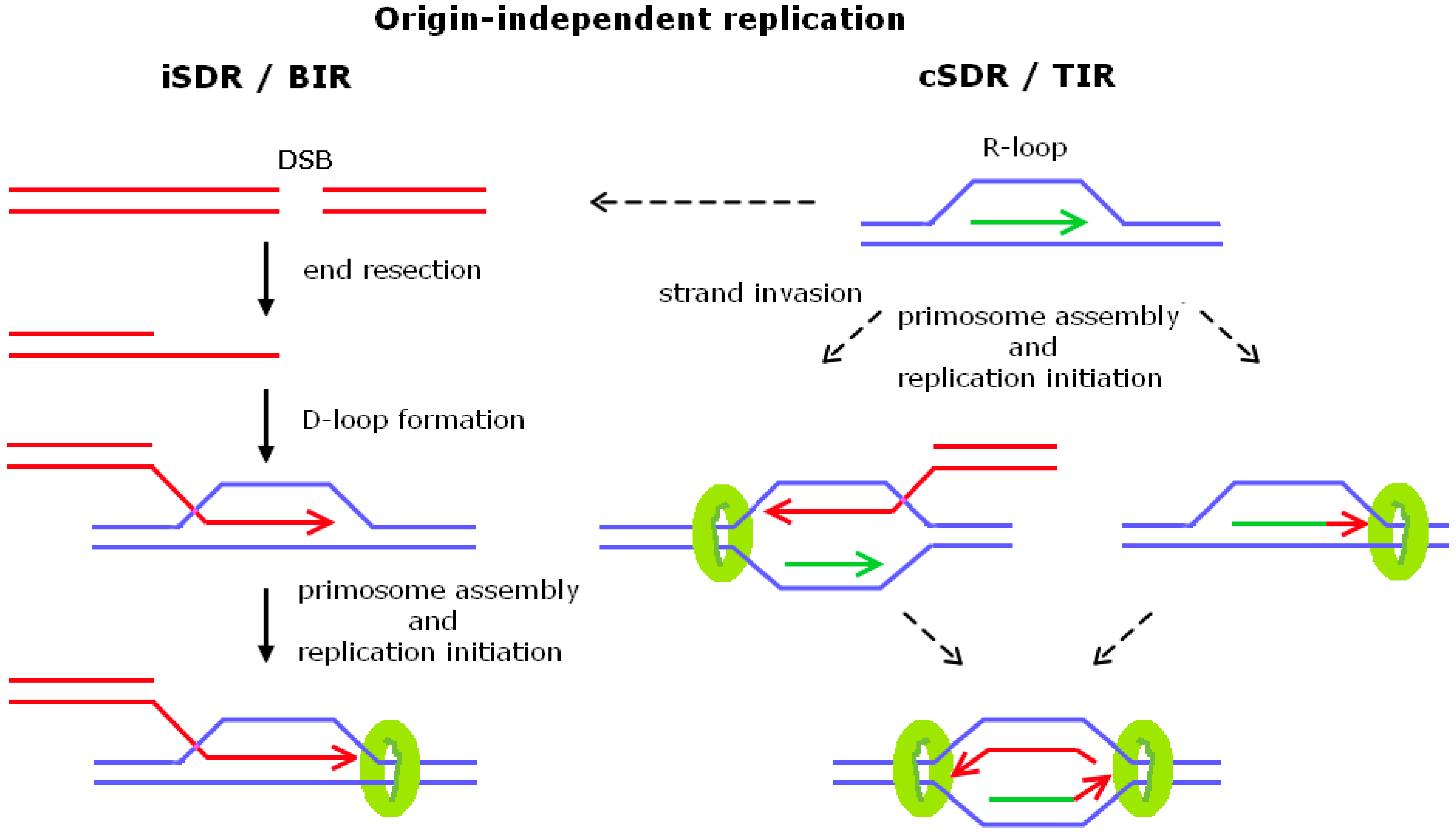
R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli | Nature Communications
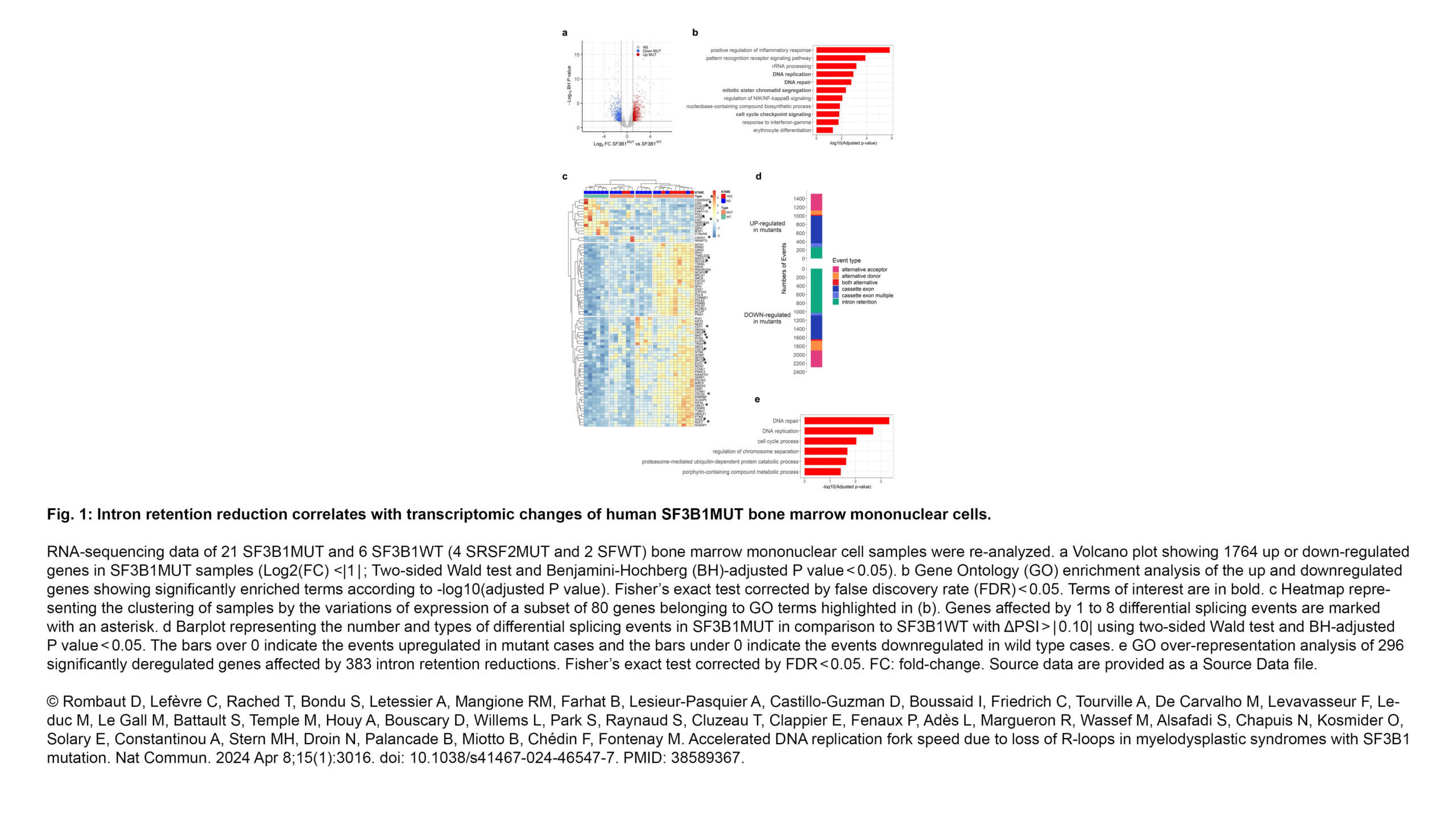
Palancade Lab - Accelerated DNA replication fork speed due to loss of R-loops in myelodysplastic syndromes with SF3B1 mutation - Institut Jacques Monod
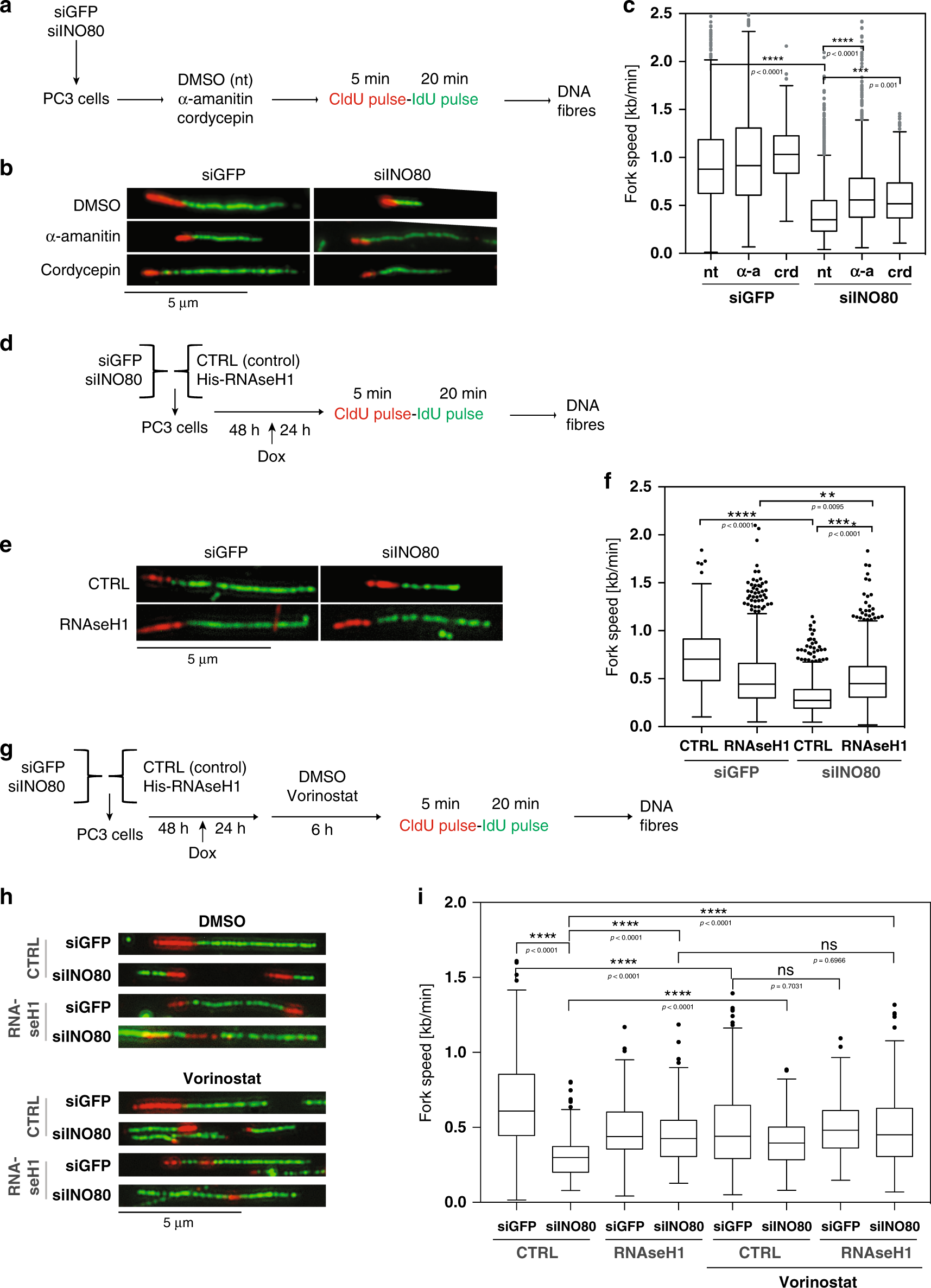
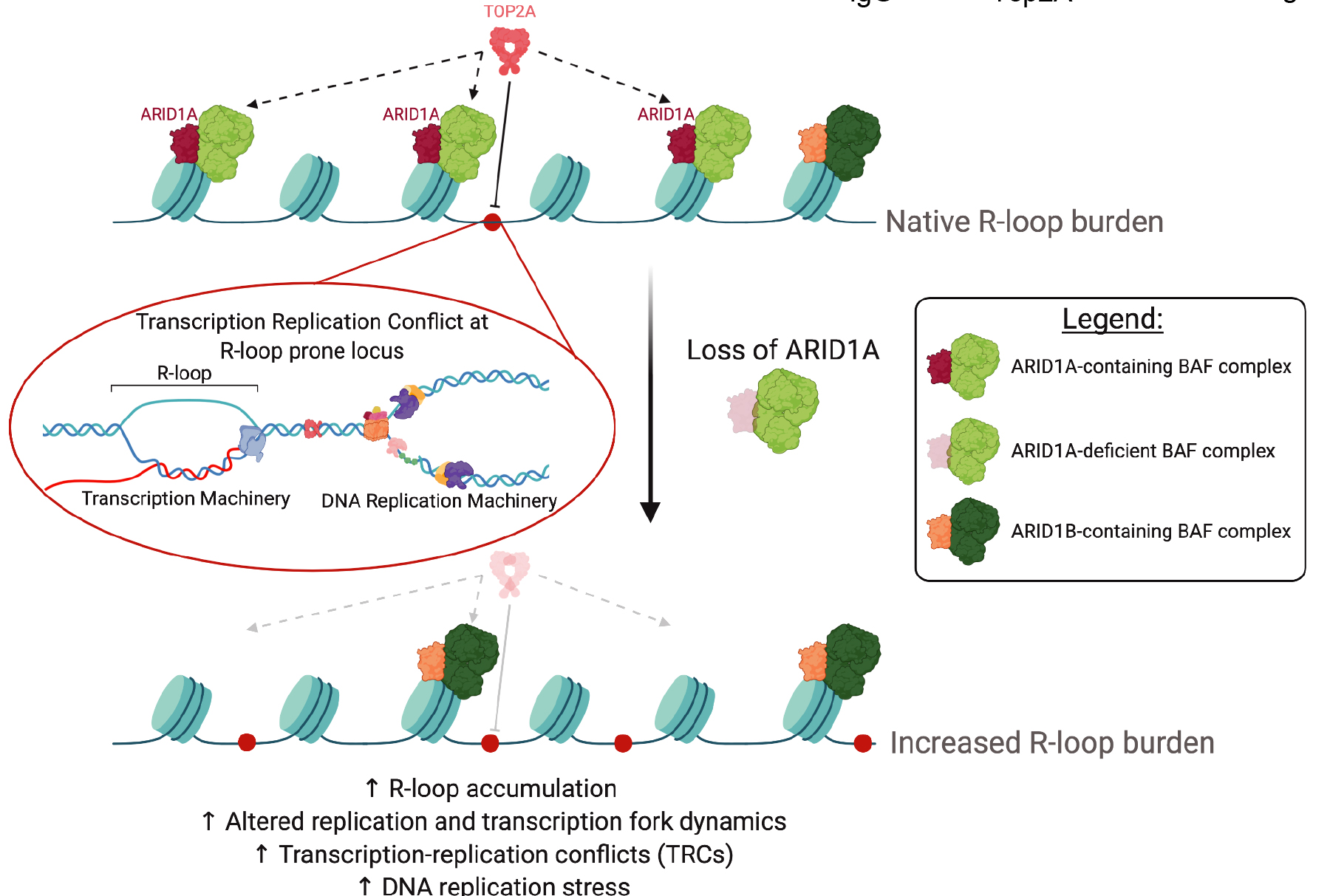
Resolution of R-loops by INO80 promotes DNA replication and maintains cancer cell proliferation and viability | Nature Communications

Replication results by collection site. Each row summarizes the effect... | Download Scientific Diagram

SciELO - Brasil - Optimum plot size and number of replications for experiments with the chickpea<sup/> Optimum plot size and number of replications for experiments with the chickpea<sup/>
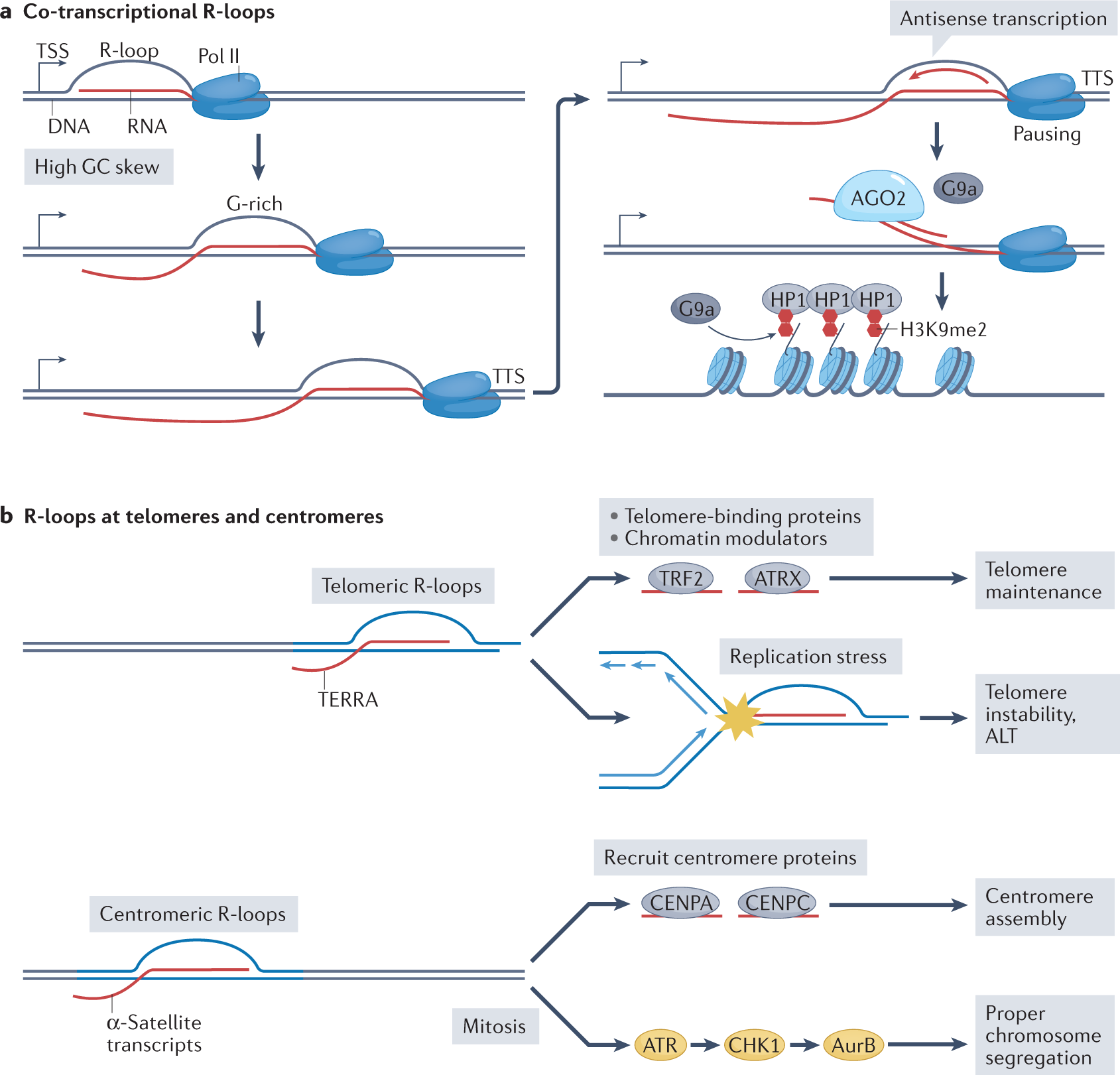
Sources, resolution and physiological relevance of R-loops and RNA–DNA hybrids | Nature Reviews Molecular Cell Biology